SpanBERT: Revolutionizing NLP with Enhanced Span Prediction – A Scientific Exploration
Introduction: In the realm of Natural Language Processing (NLP), the SpanBERT model stands as a pioneering development, offering a unique approach to text understanding. Designed to improve span prediction, SpanBERT has shown exceptional promise in various NLP tasks. In this article, we will delve into the details and underlying formulas that drive the mechanics of SpanBERT, designed specifically for the scientific community.
Understanding SpanBERT’s Architecture:
SpanBERT builds upon the transformer architecture, just like BERT, but it introduces enhancements that enable it to excel in tasks requiring span prediction, such as named entity recognition and coreference resolution.
Key Formulas and Mechanisms:
1. Self-Attention Mechanism: Like most transformer-based models, SpanBERT relies on the self-attention mechanism to weigh the importance of different words within a sequence. The self-attention formula can be represented as:
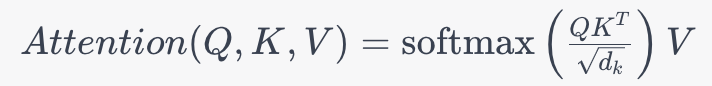
Where:
- Q represents the query vector.
- K represents the key vector.
- V represents the value vector.
- dk is the dimension of the key vector.
2. Positional Encoding: To consider the order of words in a sentence, SpanBERT employs positional encoding. This is crucial for understanding the sequence of words. The positional encoding formula is as follows:
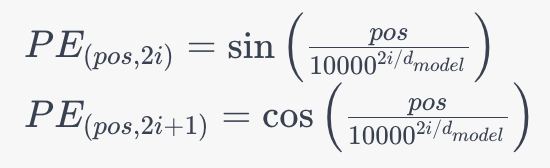
Where:
- pos represents the position of the word in the sequence.
- i represents the dimension of the positional encoding.
- dmodel is the dimension of the model’s embeddings.
3. Training Objectives: SpanBERT is fine-tuned with objectives specifically designed for tasks that require span prediction. For instance, it may use span prediction tasks where the model is trained to predict the beginning and end of spans, which are particularly useful in named entity recognition.
Applications in Scientific Context:
SpanBERT’s focus on span prediction has made it a valuable asset in the scientific community:
- Named Entity Recognition (NER): In scientific literature, identifying entities like genes, proteins, and diseases is essential. SpanBERT excels in NER tasks, aiding in the extraction of valuable information from scientific documents.
- Coreference Resolution: Scientific papers often reference entities multiple times with different terms. SpanBERT’s ability to handle coreference resolution assists in establishing connections and relationships between different mentions of the same entity.
- Information Extraction: SpanBERT contributes to information extraction from scientific texts, helping researchers glean structured knowledge from unstructured documents.
Conclusion:
In the world of Natural Language Processing, SpanBERT’s enhanced span prediction capabilities have opened new doors for scientific research and data extraction. With its meticulously designed architecture, self-attention mechanisms, and positional encoding, SpanBERT has emerged as a powerful tool for understanding and extracting valuable information from scientific texts. As the scientific community continues to embrace this technology, we can anticipate SpanBERT to be instrumental in advancing the frontiers of knowledge in various scientific domains.
